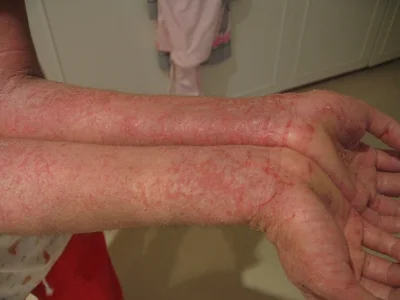
Atopic dermatitis, also known as eczema, is a skin inflammation and rash that has an enigmatic cause. There are many genetic and environmental risk factors involved, but to date the exact triggers and mechanisms that cause this autoimmune response are unknown. Importantly, a growing percentage of infants and toddlers are developing this disease, which itself is a known risk factor for other autoimmune diseases like asthma and allergies. Discovering the cause of atopic dermatitis is an important endeavor, because it may lead to a cure for a number of other diseases.
Staphylococcus aureus has long been associated with atopic dermatitis. It appears to occur at relatively high abundances in the areas of skin that are affected. Then again, S. aureus is a one of the most common skin microbiome bacteria (it is ubiquitous around the world), and it has yet to be definitively connected to the disease. In addition, mouse models for many skin diseases, including this one, do not exist or are insufficient, so controllably studying the atopic dermatitis is difficult. Recently though, a team of scientists from Japan and the NIH developed a mouse model for atopic dermatitis, and made a new discovery that showed S. aureus can indeed drive skin inflammation. They published their results in Cell immunity.
The scientists were studying how a specific genetic mutation in mice affected bones and hair follicles when they serendipitously realized that it was causing eczema in the mice after around three weeks. When they investigated the skin microbiomes of these mice, as well as normal mice, they realized that right around the time that the eczema was appearing in the mice, these mice’s skin microbiomes drastically shifted. First, a bacterium called Corynebacterium mastitidis emerged, followed by S. aureus a few weeks later, which was coincident with the presentation of the worst symptoms. Of note, species of Corynebacteria are associated with eczema in humans, much like S. aureus.
Next, the researchers then performed a series of experiments by providing mice with antibiotics in an effort to combat the dysbiosis. When newborn mice with the genetic modification were treated with antibiotics they never developed eczema at all. Moreover, genetically modified mice that were in the midst of the rash that were treated with antibiotics had their eczema subside soon after. In addition, genetically modified mice that were taken off of antibiotics had eczema emerge shortly thereafter. Strikingly, in all of the above situations changes in the skin microbiome corresponded with the disease state: a lack of S. aureus and high diversity were associated with healthy skin, and the emergence of S. aureus and a lack of diversity were associated with the disease. Finally, when S. aureus was inoculated onto the skin of genetically modified mice, they developed eczema rapidly.
The researchers performed a number of other experiments to try and tease out the mechanism by which S. aureus causes atopic dermatitis. Their results show that it appears a combination of genetic and environmental factors that affect the skin may be important in defining an individual’s risk for the disease. Regardless, it appears that S. aureus is a major culprit in causing eczema, so future therapies that eradicate the bacteria, or at least decrease its abundance should be considered.