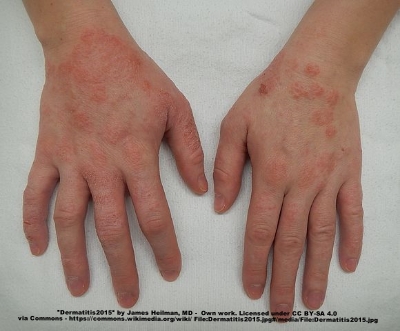
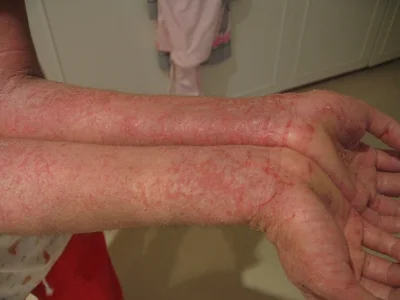
A moderate case of hand dermatitis
Atopic dermatitis, otherwise known as eczema, is an inflammatory autoimmune response of the skin. Today in the United States it affects around 25% of children, and as many as 3% of adults, with its incidences increasing each year. Like many other allergies, the microbiome is now being implicated in the cause of this disease. A few months back evidence was published linking atopic dermatitis to the skin bacteria Staphylococcus aureus. Other work, however, has shown that the gut microbiome may be critically important to this disease as well, especially because gut bacteria are more likely to control and elicit certain inflammatory responses seen in dermatitis, such as the release of specific cytokines. A group of Korea recently compared the gut bacteria in atopic dermatitis patients and healthy controls and identified a specific organism that may be important to the disease. They published their results last week in the Journal of Allergy and Clinical Immunology.
The researchers measured the gut microbiomes of 132 people, including 90 of which had atopic dermatitis and were seeking medical treatment. They also measured gene expression by bacteria in the gut, and short chained fatty acids (SCFAs) in the guts of all the individuals. They discovered that one particular bacterial species was much more abundant in dermatitis patients compared to controls, Faecalibacterium prausnitzii. After, they measured SCFA production, and noted that a decrease in butyrate and propionate was directly linked with the presence of F. prausnitzii, suggesting an important link between this bug, SCFAs, and the disease state. In addition, they noted that the overall diversity of bacteria was similar in all microbiomes measured. Finally, the scientists investigated the gene expression, and observed an increase in bacteria that are capable of breaking down gut mucins, or mucous, in the guts of atopic dermatitis individuals. For example, these bugs were expressing proteins that break down fucose and N-acetylgalactosamine (GalNAc), two monosaccharides that are normally derived from mucins rather than food.
This study presents a number of differences in the gut microbiomes of individuals with an without atopic dermatitis. The scientists suggest that an important species associated with this disease may be F. prausnitzii, and perhaps it may even be influencing the disease through a lack of SCFA production, and the breakdown of gut mucins. Atopic dermatitis is a complex disease, and certainly cannot be explained by the presence of an individual bug. However, this paper does support the notion that diseased individuals, who present rashes on their skin, may have disruptions to gut, and that changes in the gut microenvironment create a niche for specific bacteria to grow. This, in turn, may inform new therapeutic strategies that target the gut microbiome, rather than topical treatments.