Breastmilk is critically important to developing a healthy infant gut microbiome. The combination of oligosaccharides found in breastmilk are not found in any other individual food, and are intended to cultivate healthy bacteria in the gut. Besides breast milk, really the only other fluid an infant consumes is his or her own saliva, but thus far not much is known about the role this saliva plays in culturing the proper microbiota. A team of researchers from Australia recently studied how a mother’s breastmilk directly interacts with her infant’s saliva. They discovered that when combined, saliva and breast milk produce specific molecules that inhibit the growth of some bacteria, but support the growth of others. They published their results in the journal PLoS ONE.
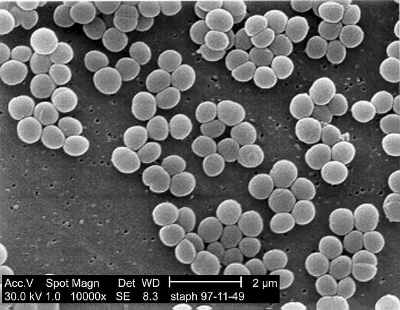
The researchers measured the molecular components of saliva in 77 adults and 60 infants. They noticed some stark differences between the two types of saliva, including markedly higher levels of salivary hypoxanthine and xanthine. Hypoxanthine and xanthine are both substrates for a protein called xanthine oxidase (XO), which reacts with them to form hydrogen peroxide (H2O2). One of the places XO is predominantly found is in human breast milk, which led the researchers to hypothesize that xanthine and hypoxanthine in infant saliva reacts with XO in breastmilk to form H2O2. Hydrogen peroxide is a reactive oxygen species (ROS) that can kill bacteria. The scientists believe that infant saliva reacts with breast milk to form hydrogen peroxide at high enough levels to kill opportunistic pathogens, but allow others to grow. In order to test their hypothesis, the researchers combined breast milk and infant saliva and attempted to culture the pathogen Staphylococcus aureus, along with gut commensal bacteria Lactobacillus plantarum, and Escherichia coli. They found that the mixture created concentrations of hydrogen peroxide that killed the S. aureus but allowed the commensals to grow.
Overall this paper showed that infant saliva can combine with breast milk to form physiologically relevant concentrations of hydrogen peroxide. The hydrogen peroxide may in fact select for the growth of specific bacteria in the mouth and gut, and lead to the development of a healthy microbiome. Interestingly, pasteurized cow’s milk and infant formula did not contain XO, the enzyme necessary to create the hydrogen peroxide, adding another reason why there is no true substitute for breast milk.