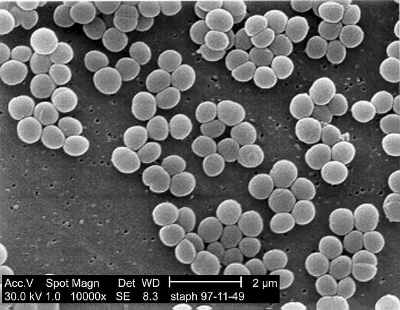
Electron micrography of Staphylococcus aureus.
The nasal microbiome remains largely unstudied despite its potential importance to many diseases, such as rhinosinusitis, allergies, and staph infection (incuding MRSA). Staphylococcus aureus is probably the most well-known nasal resident, but simple questions, such as which species of bacteria are most prevalent in the nose, are still not answered. Understanding all the residents of the nasal microbiome, the influence of our genetics and the environment on defining their populations, and the influence each one has on others may be critically important to preventing diseases such as staph infection, and more research is needed. Fortunately, a new study out of Johns Hopkins that investigated sets of twins shed light on many of these questions, and was published in Science Advances last week.
The scientists sequenced the nasal microbiomes of 46 identical and 43 fraternal pairs of twins. First, thy learned that these people’s nasal microbiomes could be classified into 7 different phenotypes or community state types (CST) which broadly described their nasal microbiomes. These 7 types are defined by their most abundant bacteria, and are as follows: CST1 – S. aureus, CST2 - Escherichia spp., Proteus spp., and Klebsiella spp., CST3 - Staphylococcus epidermidis, CST4 - Propionibacterium spp., CST5 - Corynebacterium spp., CST6 - Moraxella spp., and CST7 - Dolosigranulum spp. The most common CTS was CTS4 with 29% of the sampled population having that CTS, whereas CTS4 was the least popular, coming in at 6% of the individuals tested. The researchers noted that many of these bacteria, such as Proteus, were not considered to be important to the nasal microbiome at all, so their dominance in some noses was surprising. The scientists learned that genetics plays nearly no role in the microbiome community composition, but does influence the overall microbiome population. In addition, gender influenced the overall population, with women having about half as many total bacteria in their noses as men.
With regards to S. aureus, while it existed in 56% of the individuals studied, it was associated with other bacterial. For example, the researchers discovered that Dolosigranulum, and Propionibacterium granulosum were negatively correlated to the existence of S. aureus, whereas S. epidermidis was positively correlated with S. aureus abundance. This lends itself to the idea that specific bacteria can create colonization resistance against S. aureus, and thus could be used to prevent the disease. The researchers suggest a probiotic should be tested for its therapeutic value in preventing S. aureus colonization, and hopefully they move forward with those trials.