I recently had the pleasure of hearing a lecture by Eric Alm, an associate professor at MIT. During the lecture he described a number of studies his group has performed, and I would like to share some of his conclusions.
In his first study he and his student sampled their own microbiome every day over the course of one year, while cataloging every minute detail of their lives over the same time frame. They were investigating what activities have real effects on the microbiome, and as they discovered, unsurprisingly, what one eats is the most important. They found that the amount of fiber in the diet perturbed the microbiome most, in addition to things like orange juice, yogurt, fruits, and soup. They also discovered that after flossing, a certain bacteria from their mouth, would show up in their stool. More importantly, they confirmed that the microbiome is robust, and rebounds after drastic changes like vacationing in foreign countries. In addition, they learned that perturbations occur within 24 hours. Most of the results of the study can be found here.
In another study, Eric discovered that the microbiome is a hotbed for horizontal gene transfer. With so many genetically different bacteria living and evolving in close quarters there has been a great amount of genes passed around. He also discovered that the microbiome of farm animals (which are given antibiotics to gain weight), develop antibiotic resistance, which is then transferred to our own guts' microbiomes through this lateral gene transfer. The results from this study are published here.
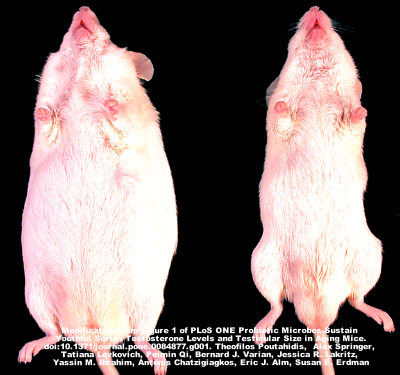
Another study focused on mice that were fed Lactobacillus in their drinking water versus those control mice that were not. The mice that were given the probiotic were skinnier than control mice, and had shinier coats and healthier skin. They then discovered that it was alterations to the immune system, rather than the Lactobacillus themselves, that were causing these changes. These results are not yet published.
Finally, Eric talked about his stool bank, OpenBiome, which we previously discussed in a separate blog post. OpenBiome is dristributing fecal material to be used in fecal microbiota transplants (FMTs). We have talked about FMTs in this blog extensively, and even touched on some studies that showed mice lose weight, or become less stressed, when given the microbiome of a healthy donor. I asked Eric if any additional phenotypes were being transferred with FMTs in humans. He mentioned in one case a patient with alopecia suddenly grew hair after the FMT, but eventually lost the hair again. In another case he mentioned a skinny woman that became obese after treatment with FMT. We will leave the reader to decide how these things play into the overall ethical controversy surrounding FMTs.